BIOSENSORS, MOTOR PROTEINS AND MORE
MARTIN WEBB
SCIENCE
Home > Biosensors and probes > Biosensors
REAGENTLESS BIOSENSORS
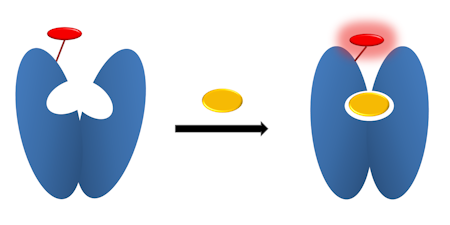
OVERVIEW
My lab has developed fluorescent reagentless biosensors based on engineered protein-fluorophore, to probe for various products of the ATPases and GTPase reactions, inorganic phosphate, nucleoside diphosphate and single-stranded DNA.
These involve placing a fluorophore as reporter at single amino acids positions on the surface of the protein scaffold, such that the fluorophore senses the ligand binding.
These have advantages that ligand binding to a protein can be highly specific, as well as rapid and tight. Using a protein as sensor enables modification of binding properties through mutation (Ref 122)
Some of these biosensors are available commercially. Others can be prepared from available plasmids and commercial fluorescent labels: see the panel, below, on "Availability". Many have been used in other labs and by Pharma.
In the brief descriptions here, main publications are referred to by number in the publication list on this website.
I would like to acknowledge and thank all the people from my lab and elsewhere, who contributed to this work. Their names are on the cited publications.
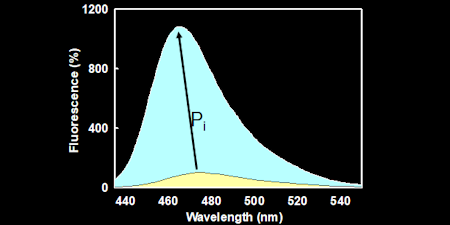
Pi - INORGANIC PHOSPHATE
Based on a bacterial, periplasmic phosphate binding protein, this gives a sensitive (Tens of nanomolar) and rapid (milliseconds) method for real-time assays of Pi formation.
It has been used for single-turnover, steady state and high-throughput applications in many labs. (General ref: 482, 102, 104)
There are three variants:
MDCC-PBP: This is the simplest to apply and uses a diethylaminocoumarin fluorophore to give ~7-fold fluorescence increase on Pi binding. (Key ref: 48, 60)
Rho-PBP: The rhodamine fluorescence gives more sensitivity and a lot more photostability in applications in which high light intensity is used over extended time. (Key ref: 89)
Rho-wPBP: This has a mutation to weaken binding and so extend the range of Pi concentrations, while using substoichiometric amounts of biosensor.(Key ref: 125).
Phosphate mop: Use of this may be required to reduce Pi contamination. See the separate panel, below.
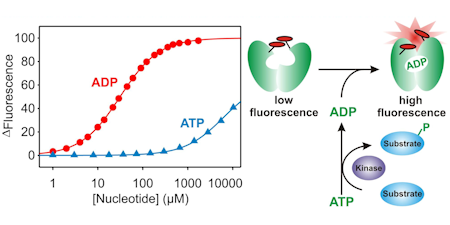
ADP and GDP
These biosensors are based on the "bacterial actin", ParM. As with the other biosensors on this page, fluorophores are located at specific sites on the surface to detect ligand binding.
There are several variants:
MDCC-ParM: This uses the fluorescence of diethylaminocoumarin and there is ~4-fold increase in fluorescence when ADP binds. There is 60-fold selectivity over ATP and this binds effectively only ADP, not other natural NDPs. The sensitivity is sub-micromolar. (Key ref: 101)
Rho-ParM: This has a weaker binding of ADP, so can measure in the tens of micromolar. It also binds GDP, so can be used to measure GTP hydrolysis, for example. (Key ref: 105, 111)
Weak-binding: We have developed some weak-binding variants that may aid ADP measurements at cellular concentrations upto hundreds of micromolar. (Currently unpublished, so please enquire about availability)
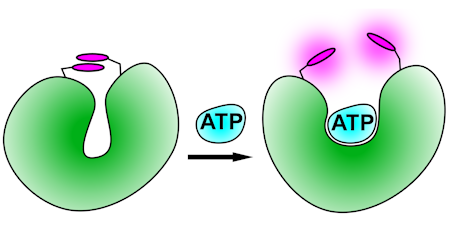
ATP
These biosensors use a malonyl-coenzyme A synthetase as scaffold, making use of the selectivity of ATP over ADP. They are adducts with rhodamines, such that ATP binding and its associated protein conformation change disrupts the stacking of two rhodamines.
In the development of these, a more structured search was made for a suitable scaffold protein, based on both binding properties and known structural changes.
Several variants have been developed that have measurable ATP ranges from sub-micromolar to >100 uM.
(126, 127)
NDP/NTP RATIO
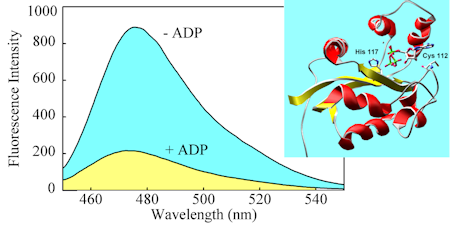
This adduct of a nucleosidediphosphate kinase with a diethylaminocoumarin senses the ratio of ADP/ATP, and is not selective for nucleoside. So it can also be used for GDP/GTP etc.
The response of this biosensor depends on the degree of phosphorylation of the kinase, giving ~4-fold change in fluorescence.
Because it does not DIRECTLY depend on binding, the biosensor responds over a wide range of nucleotide concentrations.
SINGLE-STRANDED DNA
This biosensor is based on the single-stranded DNA-binding protein (SSB) from E. coli or Plasmodium falciparum, with each subunit labeled by a single fluorophore.
DCC-SSB: This has ~7-fold fluorescence increase on binding 65-base lengths of ssDNA. It has been used to measure single-turnover helicase activity. (Key ref: 95, 103)
Cy3B-SSB: While not a biosensor giving a fluorescence change on DNA binding, this high-fluorescence probe has been used to measure ssDNA formation due to helicase unwinding in single-molecule TIRF measurements. (Key ref: 106, 110, 120, 132)
Plasmodium falciparum SSB: An alternative is DCC-SSB from Plasmodium, which has some advantages including simpler binding (so simpler interpretation) and larger signal. (131)
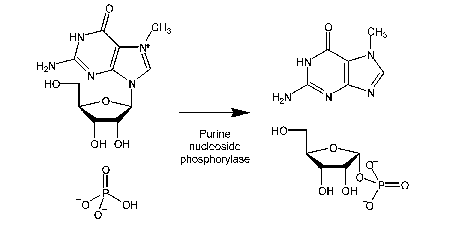
PHOSPHATE MOP
Not a new probe, but an important development for use, particularly with the Pi biosensors for high sensitivity measurements.
Pi is a ubiquitous contamination, in buffers, biochemicals such as ATP and even stuck on glassware.
This method chemically sequesters Pi, particularly prior to using the Pi biosensor (Key ref: 63)
AVAILABILITY
Most plasmids for the protein scaffolds are available from Addgene. This site also has protocols for preparation.The Pi biosensor, MDCC-PBP, may be commercially available.
The main fluorescent labels (MDCC, PBP, IATR are commercially available. Contact me for any guidance on sources and suitability.
The components for the Phosphate mop are commercially available, but it is essential to use the right grade of the phosphorylase.